Search and visualization of polysaccharides containing target monosaccharides from glycan databases.
Summary
My name is Ryu Takayanagi, a second-year master's student at the University of Tokyo, currently interning at digzyme. At university, I have been conducting research related to protein phosphorylation and protein tertiary structures.
In this tech blog, I would like to introduce GlycoSearcher, a new tool we have developed as part of our R&D activities for comprehensive search and visualization of polysaccharides containing target monosaccharides.
In recent years, research and industrial utilization of polysaccharides, such as starch and dietary fiber, have become increasingly active. There is a growing demand for the development of new saccharides, and polysaccharides, in particular, are gaining attention for their high structural diversity. To meet this need, we have developed GlycoSearcher, a tool for comprehensive search of various polysaccharides.
Description formats and databases of polysaccharides
The glyco-compounds we are focusing on have already been reported in numbers exceeding hundreds of thousands and have been databased. To selectively identify polysaccharides that fit specific purposes and apply them in fields such as synthesis pathway exploration and enzyme development, a description format that facilitates computational processing and a comprehensive database are essential.
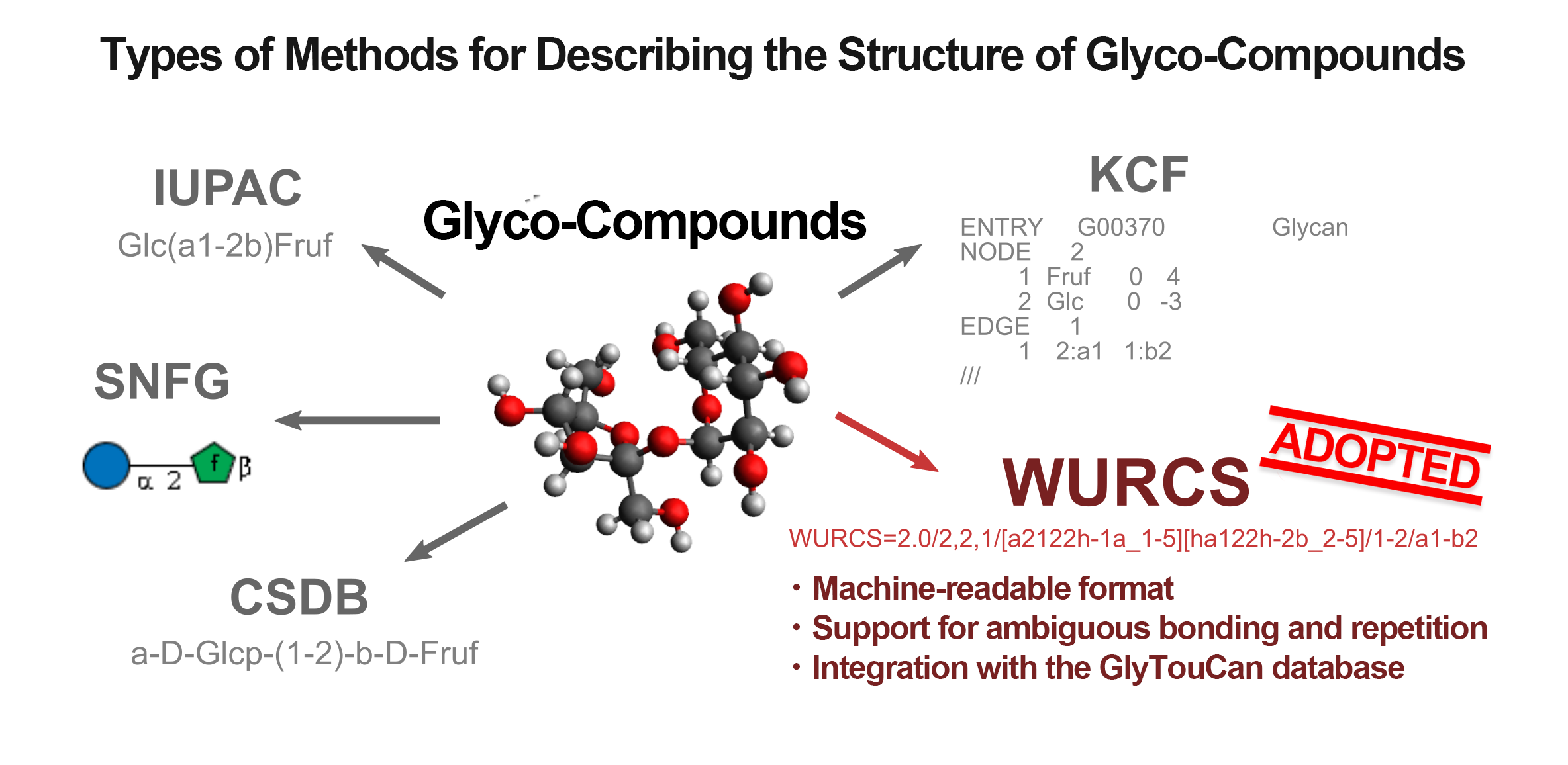
Various methods are known for describing the structure of glyco-compounds (Figure 1). Formats like SNFG and KCF excel in visualization but are not well-suited for advanced computational processing, such as structural information extraction and comparison. On the other hand, the IUPAC format offers a concise structural representation that is readable by both humans and machines, but it struggles with complex and ambiguous expressions, such as repeating units[1]. Therefore, GlycoSearcher employs the WURCS format, which is well-suited for computational processing and can represent repeating units, along with the GlyTouCan database[2], which collects glyco-compound information in the WURCS format.
(Figure 1)
Search Using GlycoSearcher
With GlycoSearcher, it is possible to extract polysaccharides that match specific criteria from a vast number of candidates. For example, you can search for polysaccharides containing particular monosaccharide units such as glucose or galactose. Additionally, it includes a filtering function that allows you to limit the monosaccharide units that make up the polysaccharides. This enables you to list polysaccharides that can be synthesized using a specific monosaccharide as a starting material and other selected sugars.
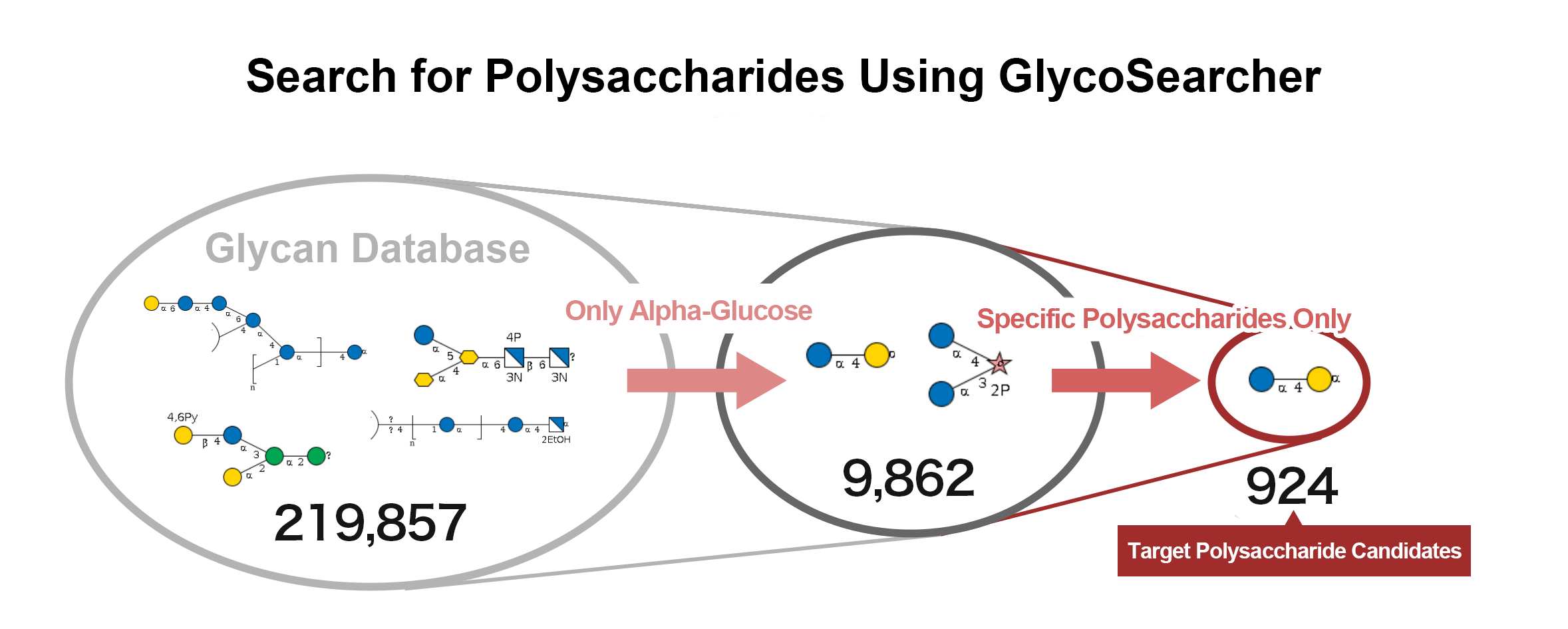
The results of a search for polysaccharides containing α-glucose are shown below (Figure 2). Out of 219,857 glycan structures, 9,862 polysaccharides containing α-glucose were identified. Further narrowing down the search to polysaccharides consisting only of glucose, galactose, and fructose reduced the number of candidates to 924.
(Figure 2)
Visualization and feature extraction of polysaccharide structures
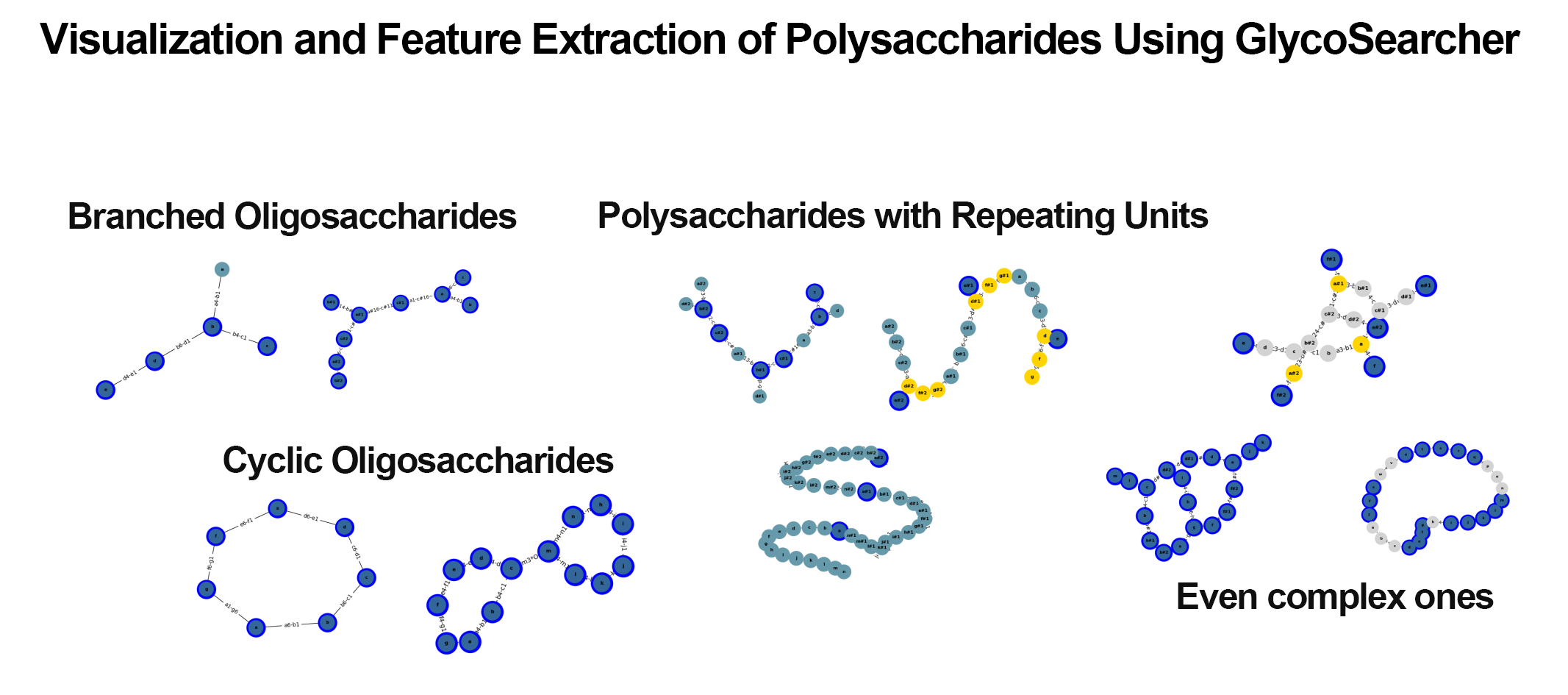
The obtained search results can be effectively visualized and utilized for subsequent applications (Figure 3). By reconstructing polysaccharides in WURCS format as graphs, it is possible to rapidly visualize thousands of search results within minutes. Additionally, for structures with ambiguous repeating units, repeating only a specific number of times allows not only the visualization of the actual structure but also facilitates further computational processing, such as structural comparisons that are challenging when ambiguous.
Since the polysaccharides in the search results are represented as graphs, feature extraction for polysaccharide structures is also possible. For example, computations can determine whether the obtained polysaccharide structures have glucose units at their termini or include specific structures (motifs). Furthermore, by integrating the hit polysaccharides with various databases such as PubChem[3], it is possible to obtain information on their common names and related enzyme information, thus providing insights into reactions involving the polysaccharides.
(Figure 3)
Conclusion
The GlycoSearcher we developed allows for comprehensive searching of target polysaccharides from the database and facilitates further computational processing. Additionally, by extracting information from the identified target polysaccharide candidates and obtaining enzyme information predicted to be involved in their synthesis, we have established a system that links to subsequent enzyme design workflows.
Acknowledgments
The development of GlycoSearcher, including acquiring knowledge about glyco-compounds, was greatly supported by Mr. Isozaki from the Business Development Department. I would like to take this opportunity to express my gratitude.
References
[1] Hosoda, M., & Kinoshita, S. (2021). "Introduction to Glycan-related Informatics." JSBi Bioinformatics Review, 2(1), 87-95.
[2] GlyTouCan. Retrieved from https://glytoucan.org/
[3] PubChem. Retrieved from https://pubchem.ncbi.nlm.nih.gov/
Practical Example of Enzyme Activity Prediction Using Structure Prediction and MD Simulation
Introduction
I am Isosaki from the Business Development Department. At our company, we are conducting enzyme activity prediction as part of our useful enzyme exploration efforts using molecular dynamics (MD) simulations. From unknown enzyme sequences, we predict their structures and subject the enzyme-ligand complexes to MD simulations. Based on the results, we calculate a proprietary digzyme score to predict enzyme activity. In this blog, we will present an example of predicting the activity of a thiolase-like enzyme, OleA, from its homologous sequences.
Materials Used for Enzyme Activity Prediction
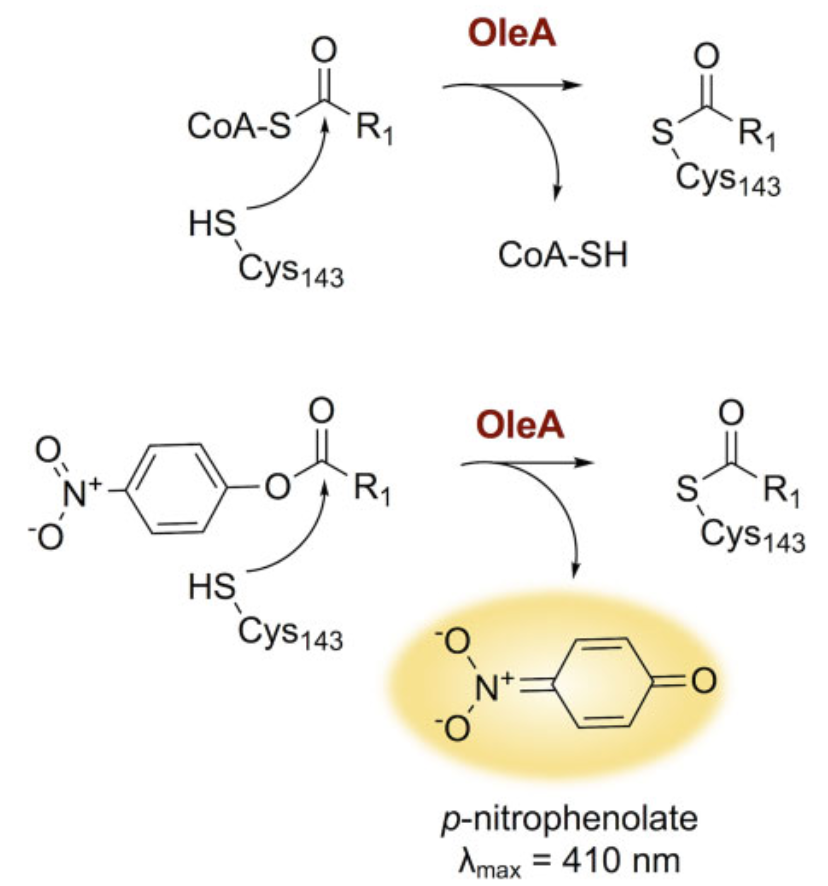
The natural substrate for OleA is acyl-CoA. The enzyme's Cys143 residue cleaves this acyl group. In investigating this activity, a p-nitrophenolate-based experimental system is used (Figure 1).
Results
We predicted whether 59 homologous OleA sequences would hydrolyze one type of p-nitrophenolate, specifically 4-nitrophenyl-hexanoate.
1. Prediction of the 3D Structure of 59 Homologous Sequences
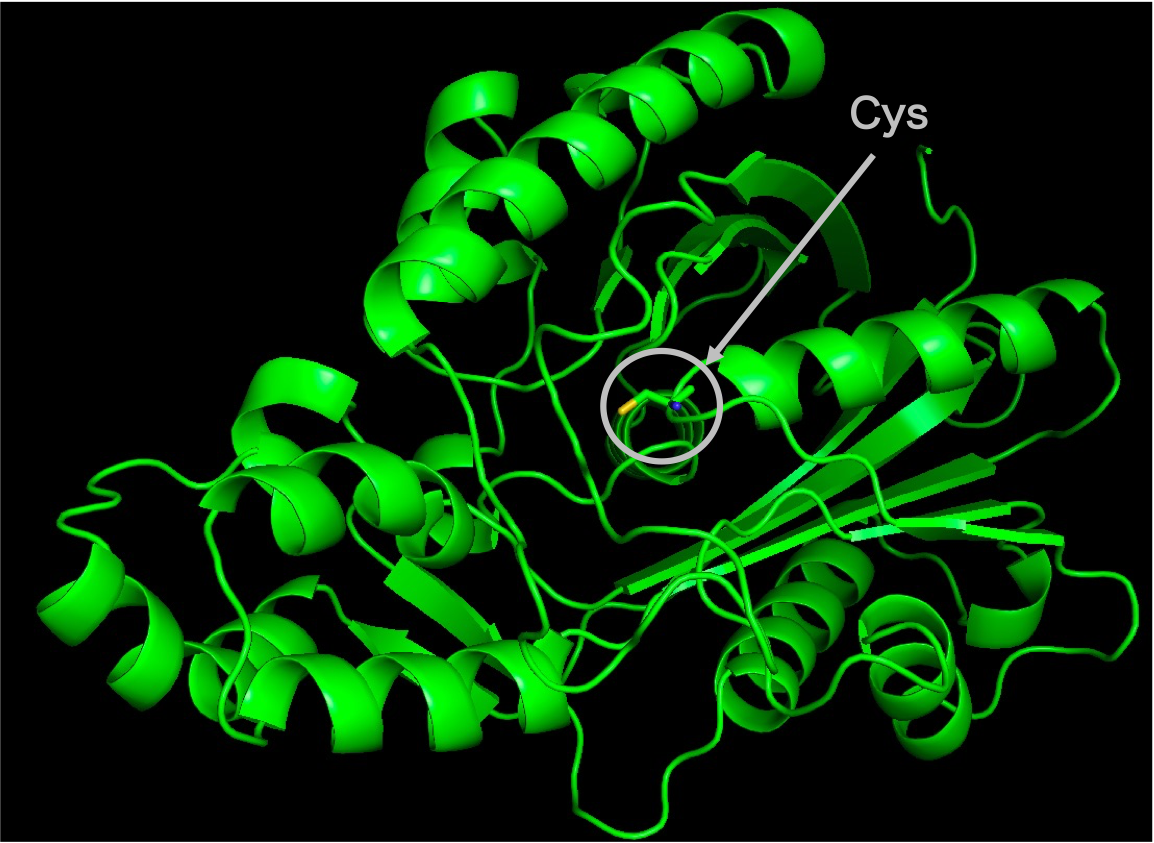
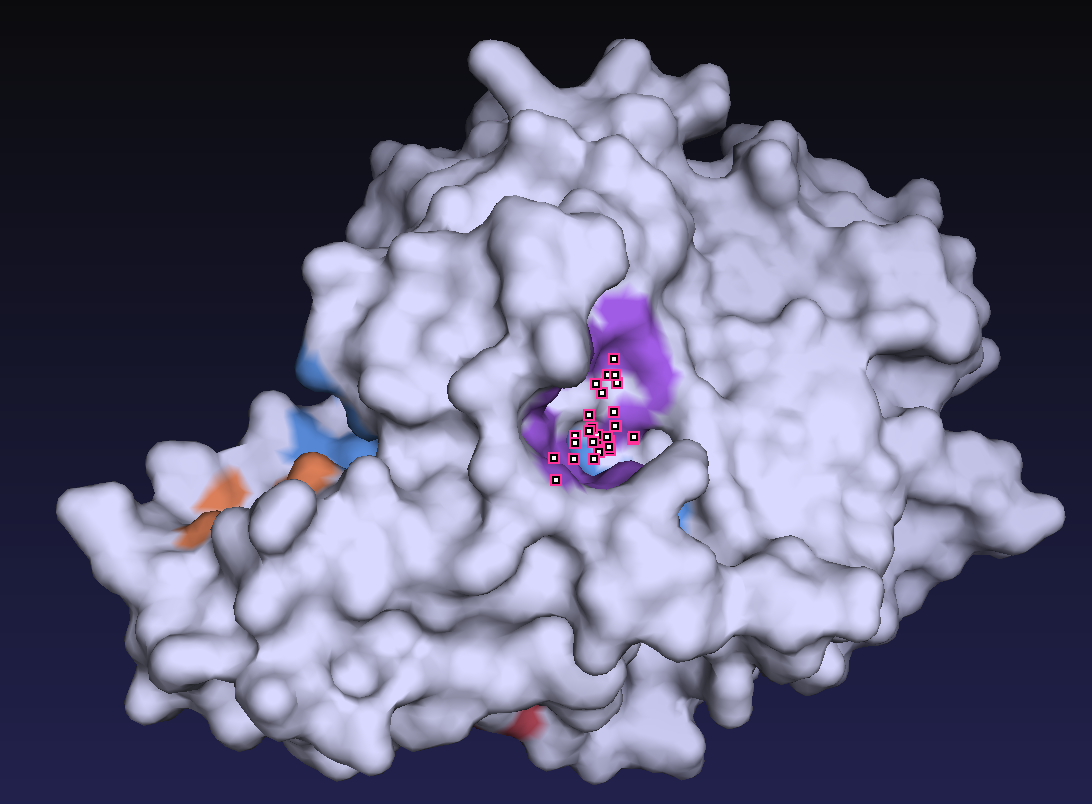
First, as all 59 homologous sequences had unknown structures, we predicted their 3D structures. From the predicted structures, we also identified the location of the active residues and the substrate-binding pocket. Figure 2 shows the predicted 3D structure of the homologous sequences and the location of the active residue, Cys. Figure 3 shows the predicted location of the substrate-binding pocket.
2. Molecular Dynamics Simulation
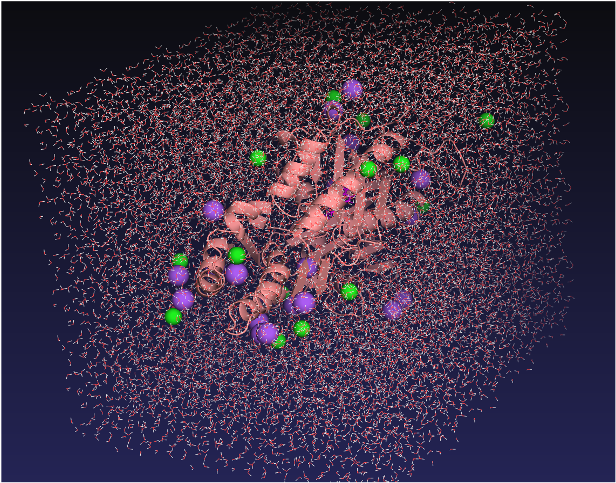
Next, we placed the enzyme-substrate complex, consisting of the enzyme and 4-nitrophenyl-hexanoate, in a system containing water molecules and ions, and ran molecular dynamics simulations (Figure 4).
3. Calculation of digzyme's Proprietary Enzyme Activity Prediction Score
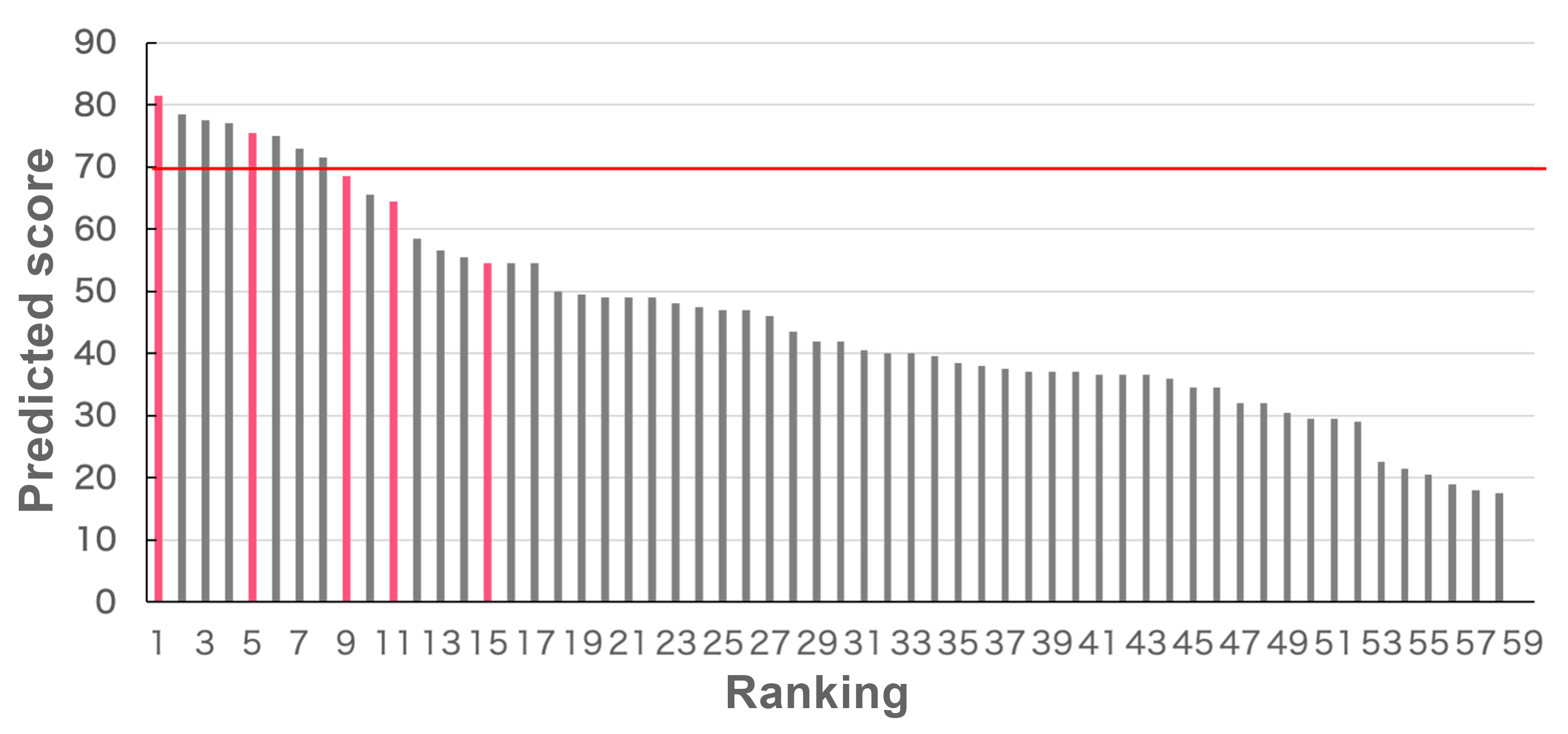
Finally, based on the results of the molecular dynamics simulations, we calculate digzyme's proprietary score. Figure 5 shows the predicted scores for all 59 sequences, sorted in descending order. Sequences with confirmed activity from experimental validation are highlighted in pink, while those without activity are shown in gray. Sequences with a score of 70 or higher were classified as active (above the red line in Figure 5). In this case, we predicted 9 sequences to be active, 3 of which were experimentally confirmed to have activity, resulting in a positive predictive value (PPV) of 0.30. The true positive rate (TPR) was 0.6, and the false positive rate (FPR) was 0.13. This indicates that inactive sequences were successfully ranked lower, while active sequences were included in the higher ranks.
Conclusion
In this blog, we demonstrated the prediction of enzyme activity using our enzyme activity prediction technology, and validated the predictions with experimental results. Typically, 5 to 10 sequences are synthesized for experimental validation. In this case, two of the top five ranked sequences were confirmed to have activity through experiments, demonstrating the practical accuracy of our enzyme activity prediction method. The dataset was specifically selected to simulate a scenario where only a small fraction of the enzyme sequences in the population exhibit activity (in this case, 5 out of 59 sequences). Since the false negative rate was kept low, we successfully predicted enzyme activity with high accuracy.
Acknowledgments
We would like to thank the following paper for providing the experimental data used in this enzyme activity prediction:
Robinson et al., (2020) Machine learning-based prediction of activity and substrate specificity for OleA enzymes in the thiolase superfamily. Synthetic Biology.
