Exploration of enzymes catalyzing unknown reactions
Introduction
This is Isozaki from the Business Development Department. At our company, we specialize in exploring enzymes that catalyze unknown reactions. By analyzing the reaction similarity to known enzymatic reactions and the sequence homology to enzymes responsible for these reactions, we can predict candidate enzyme sequences for target unknown reactions. In this blog, we present a specific example where we predicted the enzyme sequences responsible for the synthetic reaction of Islatravir, a candidate compound for HIV treatment.
Materials Used for Enzyme Exploration
We utilized data from Huffman et al., 2019. This study designed a novel synthetic pathway for Islatravir, identified enzymes catalyzing each reaction in the pathway, and validated them experimentally. The synthetic pathway of Islatravir is illustrated in Figure 1. The synthesis follows the sequence: Compound 6 → Compound 7 or 8 → Compound 5 → Compound 4 → Compound 3 + Compound 2 → Islatravir. Using our enzyme exploration technology, we predicted the enzymes responsible for each reaction in this pathway and compared them with the enzymes used in the study.

Results
First, we explored reactions similar to each of the five reactions in the synthesis pathway.
1. Similar Reaction Search
Oxidation Reaction of Starting Material 6→7 (or 8→5)
Several reactions that oxidize a hydroxyl group to an aldehyde group were extracted as similar reactions. Figure 2 shows a portion of the similar reactions with high similarity to the target reaction and their rankings. Since this reaction is not found in known metabolic pathways, multiple similar reactions were extracted.

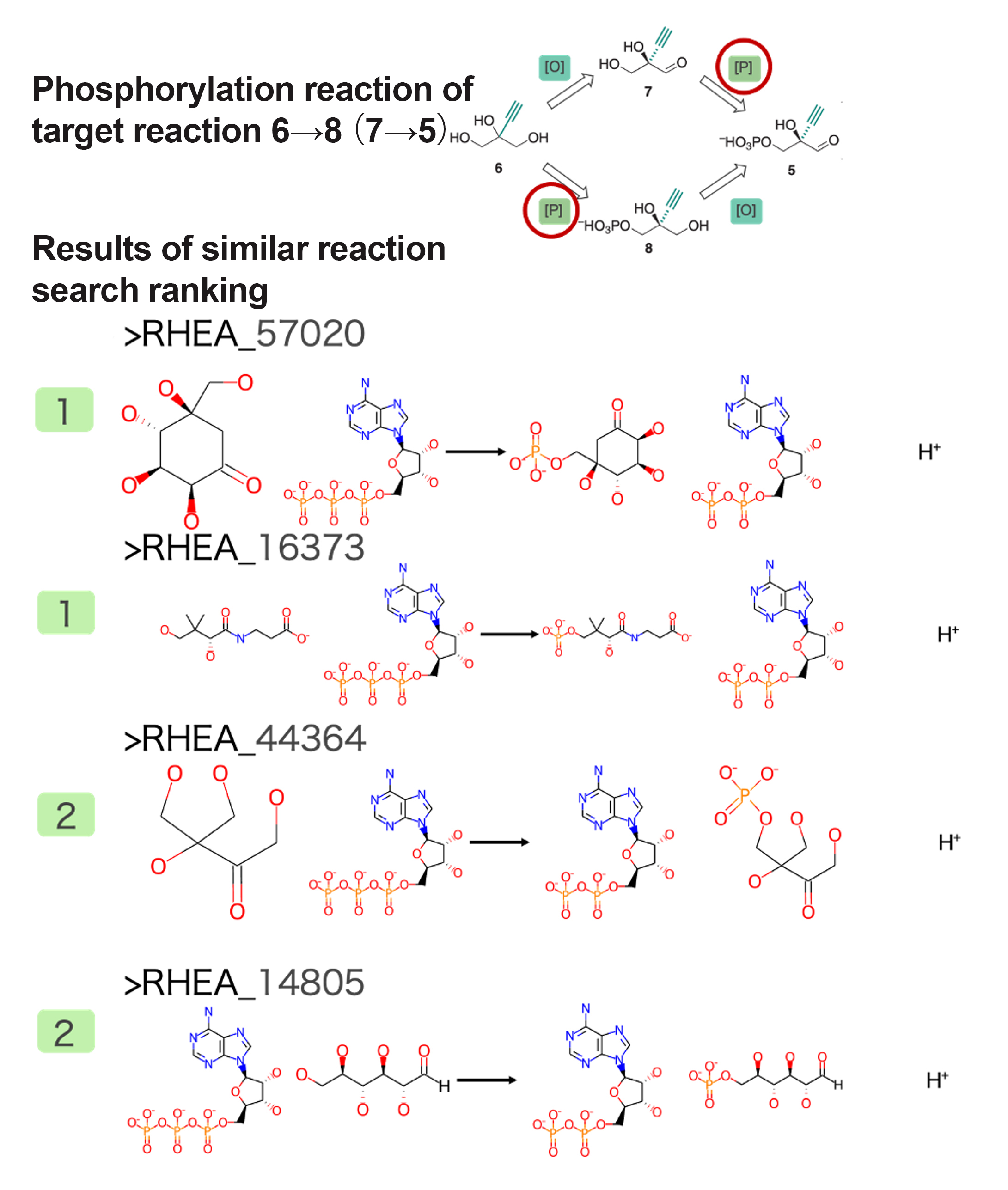
Similar Reaction for Phosphorylation Reaction of Starting Material 6→8 (or 7→5)
Several reactions that phosphorylate a hydroxyl group were extracted as similar reactions. Figure 3 shows a portion of the similar reactions with high similarity to the target reaction and their rankings. Similar to the reaction mentioned above, this reaction is not found in known metabolic pathways, so multiple similar reaction candidates were extracted.
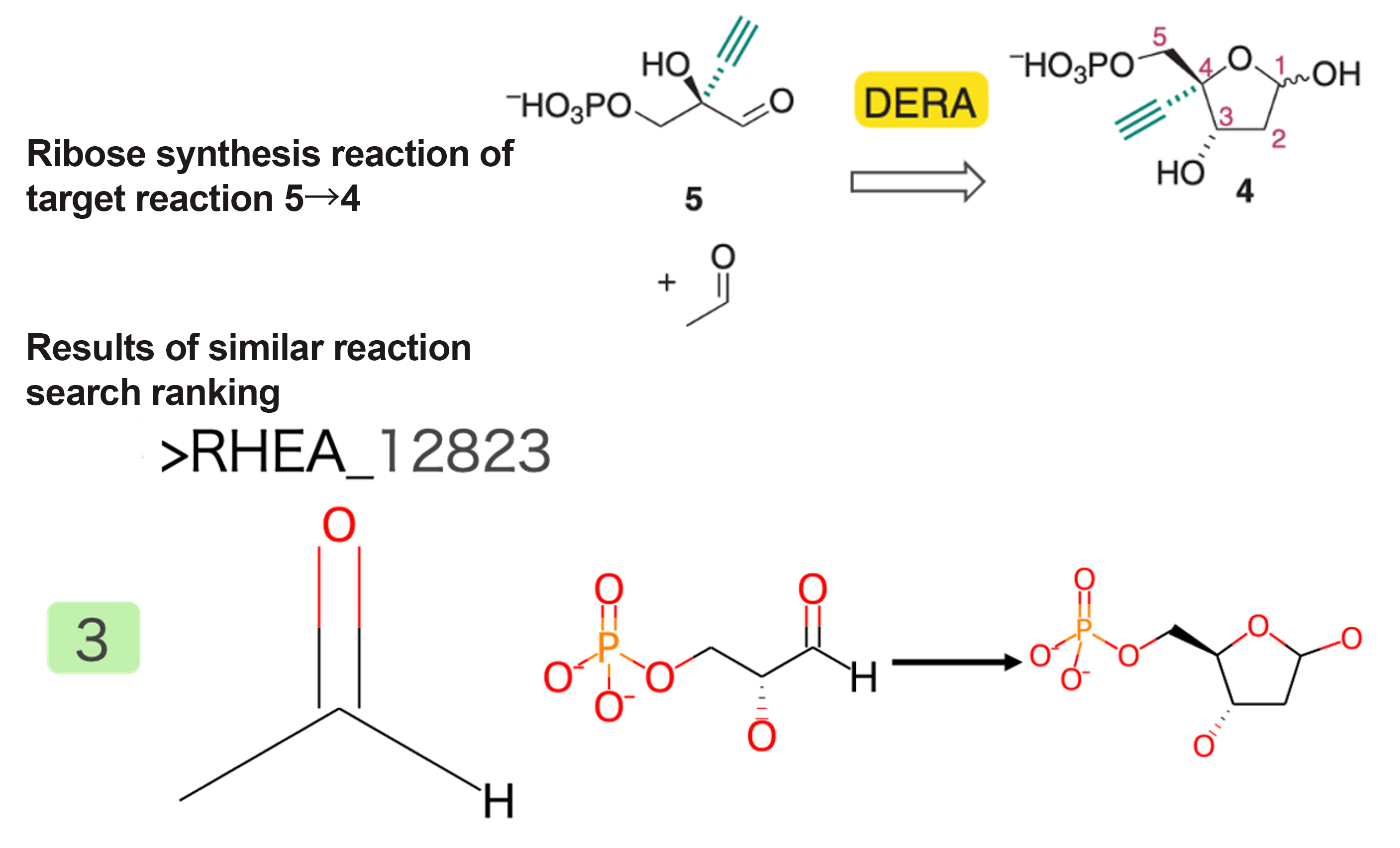
Synthesis Reaction of Intermediate 5→4 (Ribose)
A reaction that forms deoxyribose by cyclization upon the addition of acetaldehyde was extracted (Figure 4). In the paper, this reaction mimics known metabolic reactions, and therefore, a reaction that is identical except for the alkyne group was obtained as a similar reaction.
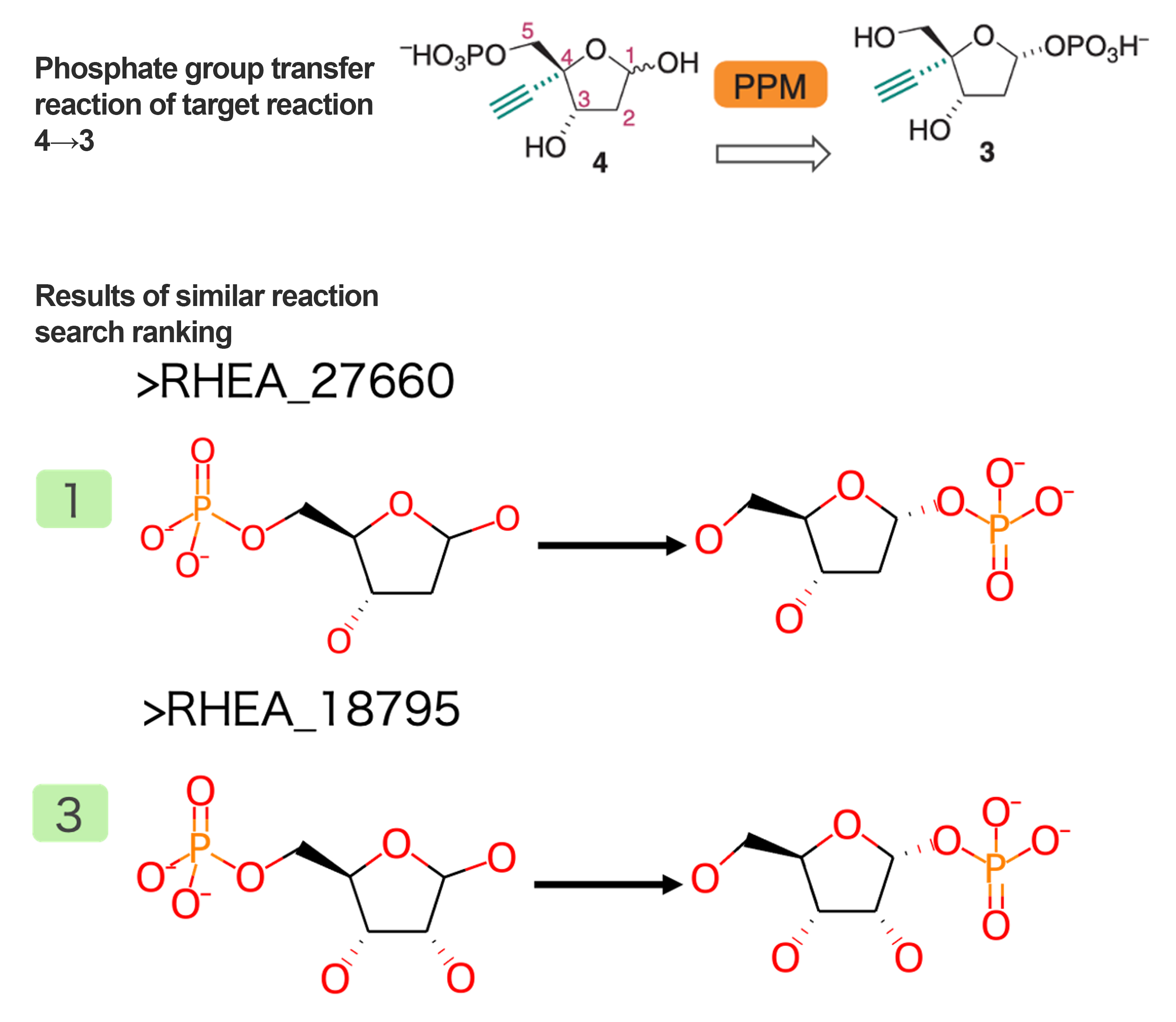
Phosphorylation Reaction of Intermediate 4→3
A reaction that transfers a phosphate group from a hydroxyalkyl group to a hydroxyl group was extracted (Figure 5). Similar to the ribose synthesis reaction mentioned above, this reaction also mimics known metabolic reactions, and therefore, a reaction that is identical except for the alkyne group was obtained as a similar reaction.
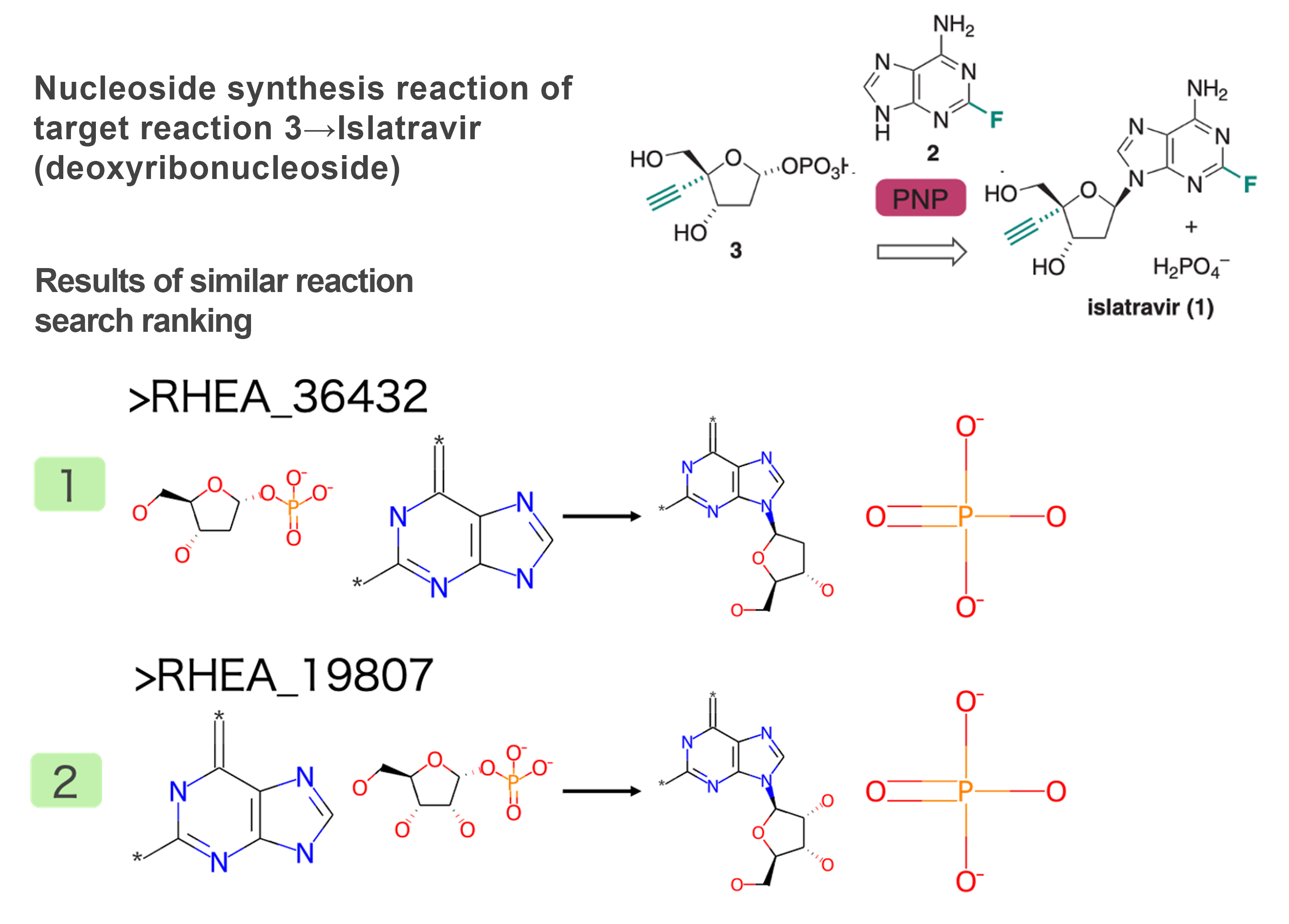
Intermediate 3 → Nucleoside Synthesis Reaction of Islatravir
A reaction that adds purine to deoxyribose was extracted (Figure 6). Similar to the above phosphate group transfer reaction, this reaction also mimics known metabolic reactions, and therefore, a reaction that is identical except for the alkyne group and fluorine was obtained as a similar reaction.
2. Search for enzymes corresponding to similar reactions
In Result 1, similar reactions for each of the five reactions were extracted. Enzyme sequences responsible for these similar reactions were extracted by taxon. For each of the five reactions, we checked whether the enzymes used in the study were included among the narrowed-down sequences. Additionally, for each of the five reactions, the enzyme sequences were further filtered using phylogenetic position screening from all taxa-derived enzyme sequences.
Extraction of enzyme sequences for similar reactions by taxon
We searched for enzymes responsible for the similar reactions in Result 1 and extracted them in three stages: from the genus Escherichia, bacteria, and all taxa. The number of sequences extracted is shown in Table 1 below. We checked whether the enzymes used in the current study were included. In four of the five reactions, the enzymes used in the study were included among the enzyme sequences extracted by our enzyme discovery technology.
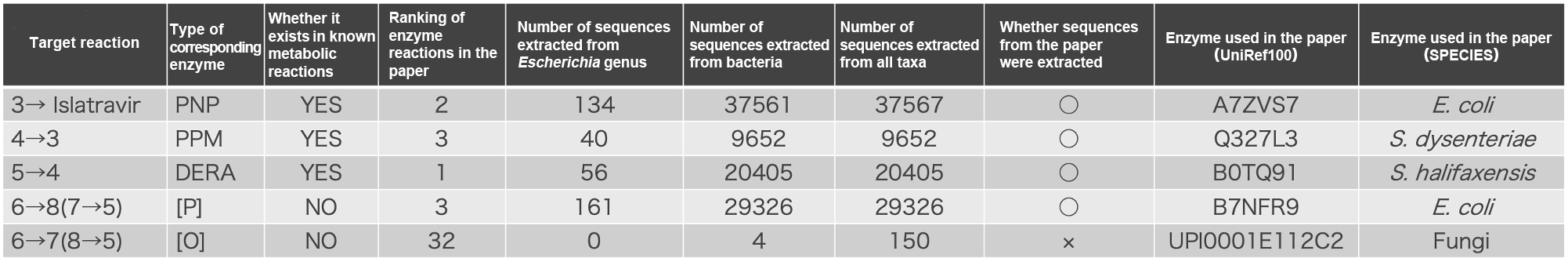
Screening based on phylogenetic position
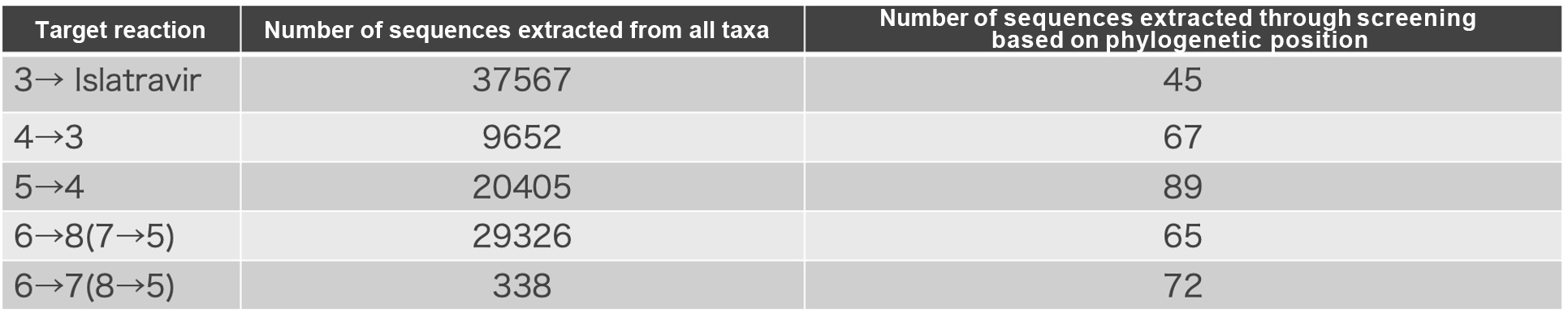
From the similar reaction enzyme sequences extracted from all taxa, we further narrow down the sequences based on their phylogenetic positions. All sequences were clustered and a phylogenetic tree was generated. From phylogenetically grouped clusters, one sequence was selected from each group. In this selection process, priority was given to sequences with high conservation across species (Table 2, Figure 7).

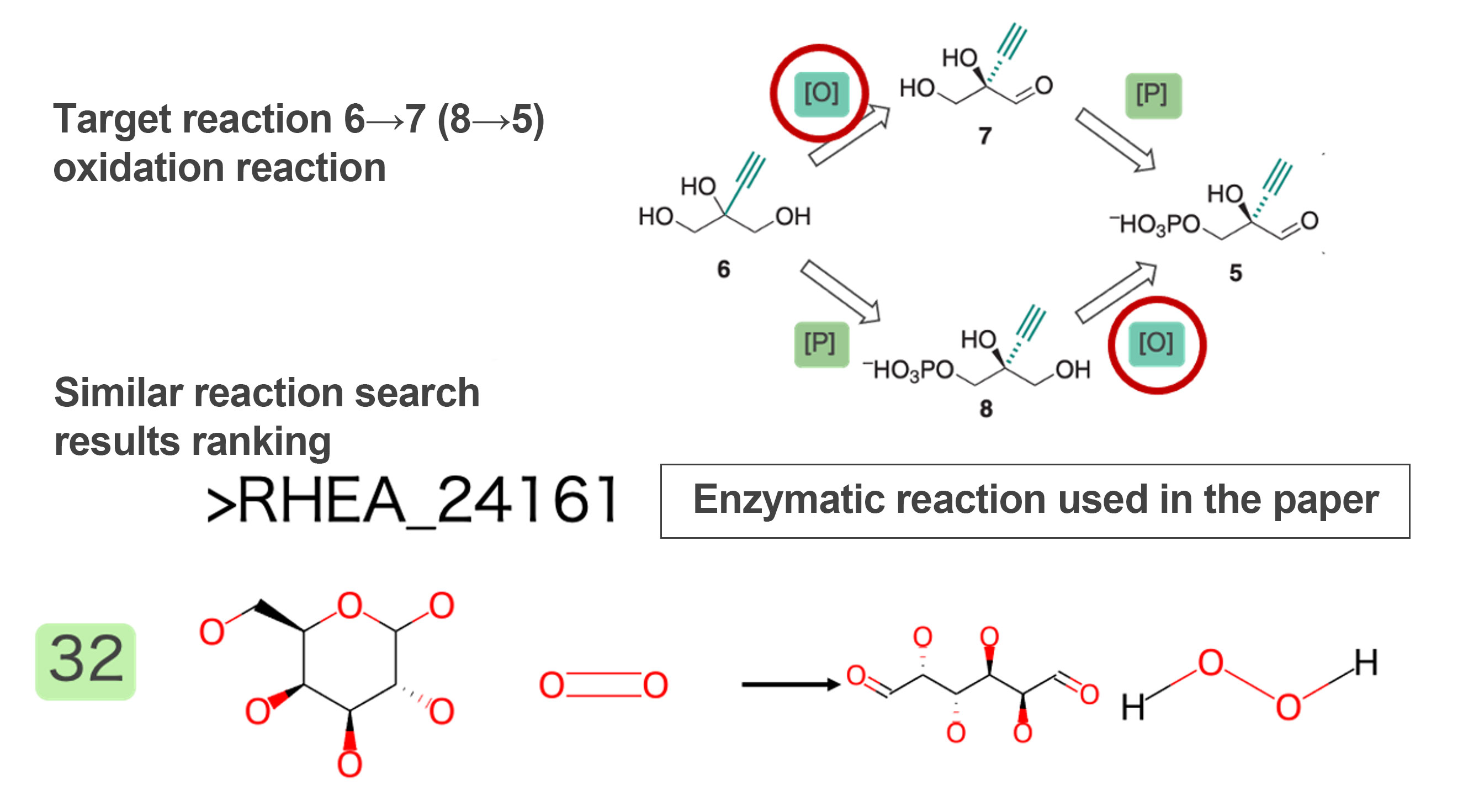
Considerations on the oxidation reaction from 6→7 (8→5)
As shown in the results above, the enzyme used for this reaction in the referenced paper was not identified in this study. One possible reason is that the target reaction, 6→7 (8→5), and the enzymatic reaction used in the paper are not sufficiently similar (Figure 8). However, the similar reactions identified through the current search also utilize O2-dependent oxidoreductases, which may catalyze the target reaction. The enzyme used in the paper for catalyzing the reaction shown in Figure 9 is estimated to be UniParc ID: UPI0001E112C2. This sequence is a member of the UniRef50 cluster, which includes sequences confirmed to catalyze RHEA_24161. However, UPI0001E112C2 itself has not been curated to confirm its catalytic activity for this reaction.
Conclusion
In this blog, we demonstrated the search for enzymes responsible for unknown reactions. We used a novel synthetic pathway for Islatravir, where we identified the enzymes that catalyze each reaction in this pathway. We attempted to find similar reaction enzymes for five unknown reactions, successfully extracting multiple similar reactions for each. In this process, we extracted sequences by arbitrary taxa and provided the number of sequences for each. For four of the reactions, we were able to extract multiple sequences that included the enzymes used in the paper. We then narrowed down the similar reaction enzymes extracted from all taxa by considering their phylogenetic positions. In standard screenings, we can further refine candidate sequences using other indicators, such as the properties of the enzymes (cellular localization, etc.) and their three-dimensional structures.
Acknowledgments
We utilized data from the following paper for the similar reaction enzyme search:
Huffman et al., (2019) Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science.
